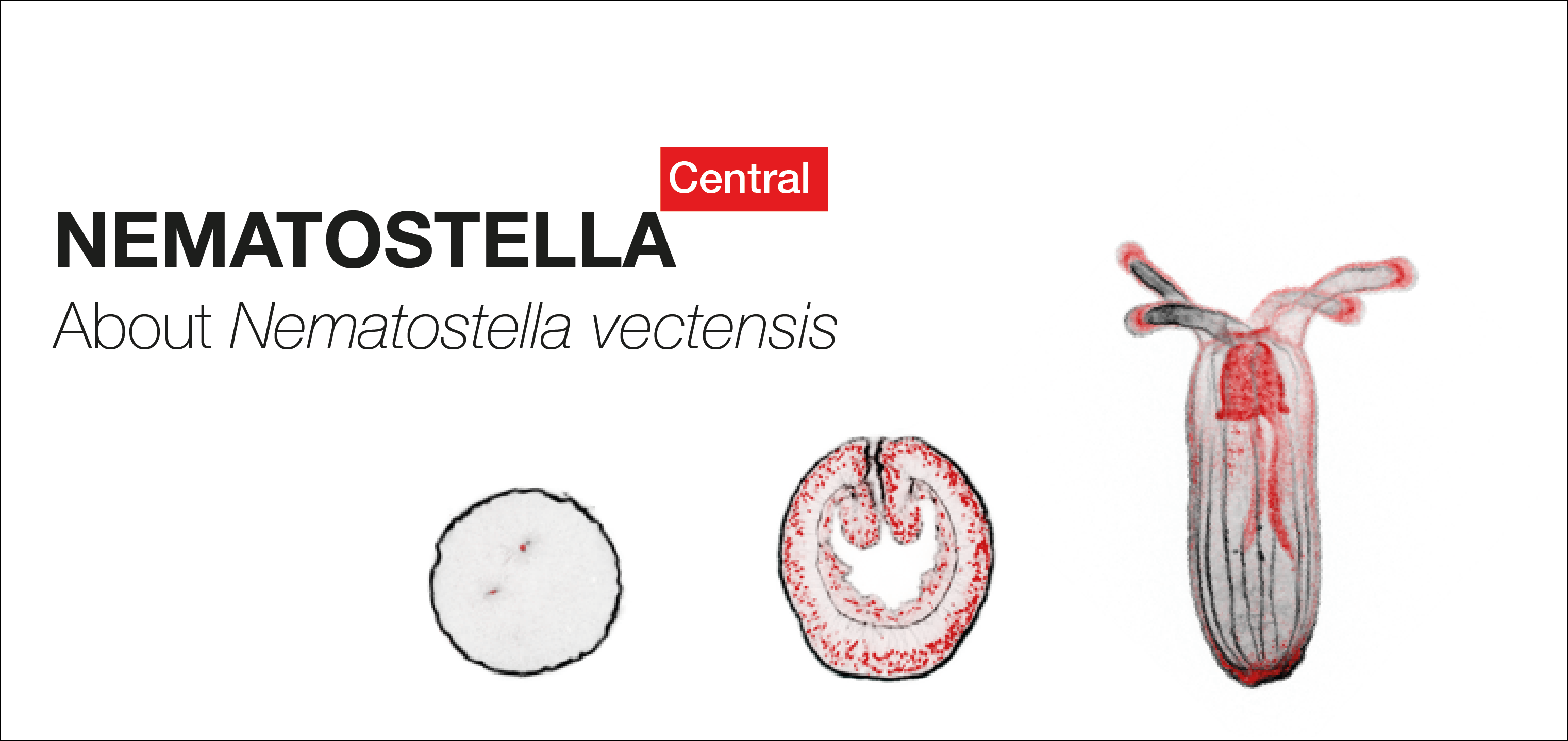
All about the starlet sea anemone
Nematostella vectensis, also called the starlet sea anemone (Cnidaria, anthozoa) lives in estuarine environments like salt marshes or brackish water pools from the Northern Atlantic Ocean. Since the 90s, this marine invertebrate has been adopted by researchers around the world to study its ecology, stress response, embryonic development or extreme regenerative capacity.
NEMATOSTELLA Central provides an overview of general information about the this research model, its embryonic development and whole body regenerative capacities, laboratories that use Nematostella for their research projects as well as links to existing ressources.
General information
Nematostella vectensis, also called the starlet sea anemone is an anthozoan cnidarian belonging to the same group of animals than stony corals. Nematostella is native to the east coast of the United States and lives in estuarine environments like salt marshes or brackish water pools. While natural populations have been located in Nova Scotia (Canada), introduced populations can be found on the West coast of the United States as well the coast of southeast England (Stephenson, 1935; Hand & Uhlinger, 1994; Sheader et al., 1997, Reitzel et al, 2013). Nematostella vectensis is a temperate sea anemone and tolerant to drastic environmental variations. In fact, the references mentioned above reported that the salinities and temperatures of the natural habitat of Nematostella can vary from 2 to 42ppt and -1.5C to 32C, respectively. Population densities can vary strongly between 120-2700 specimens/m2, according to location and season. In England, Nematostella vectensis is under threat because it is recorded from only a few restricted areas and these areas are especially vulnerable (Williams, 1991). Additional information on habitat, geographical distribution and the general biology of this sea anemone can be found at the following link: MarLIN – The Marine Life Information Network
Nematostella is translucent, colourless and a rather small sea anemone. The body column size of naturally occurring sexually mature specimen ranges between 1.5 – 4 cm in length. In the wild most of its body is buried in the sediment, with only the mouth and tentacles sticking out in the water column to catch free-living microfauna, such as copepods and larvae.Its morphology is fairly simple (see photo to the right). They are
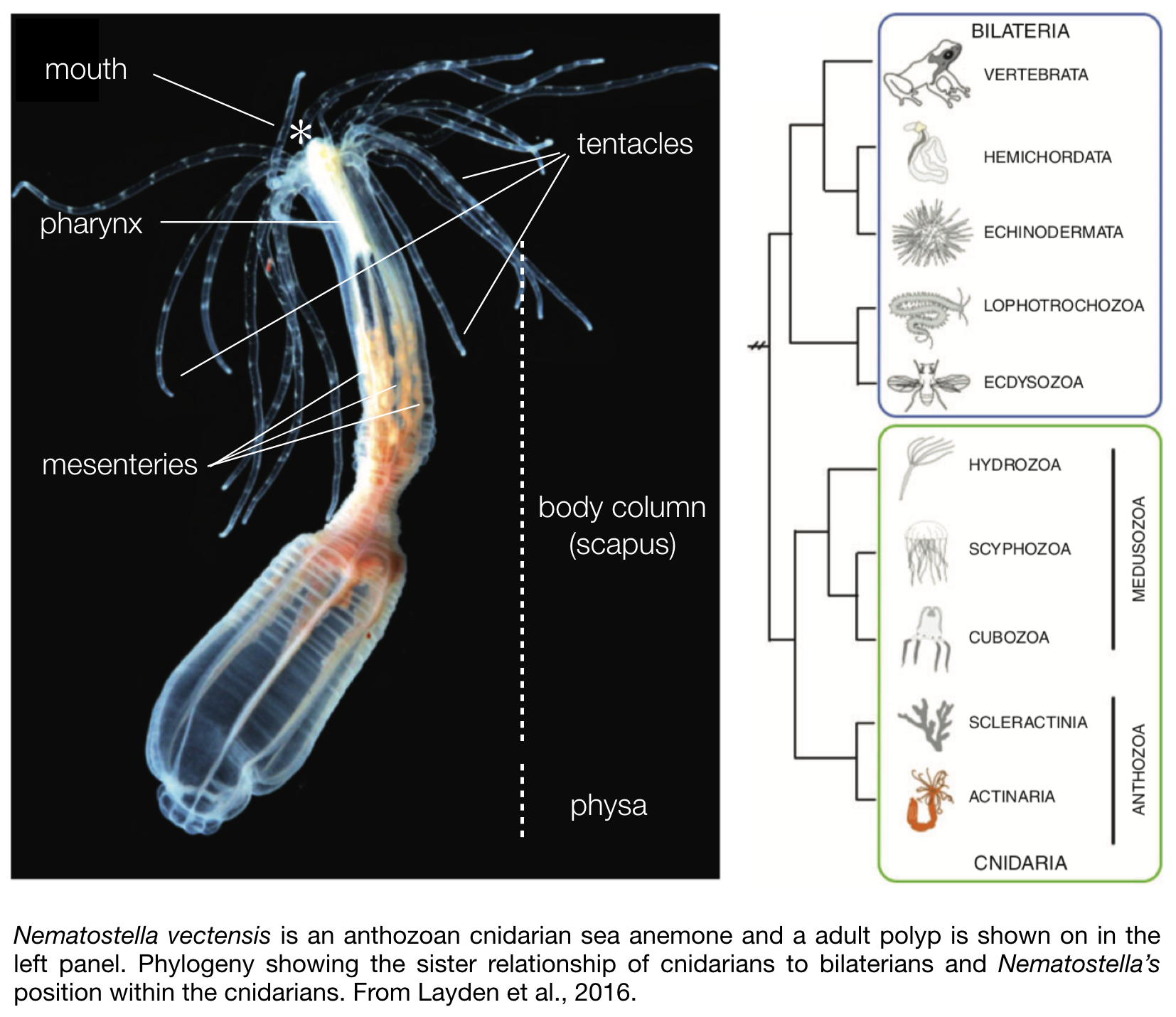
diploblastic animals, meaning that they consist of two germ layers, a bifunctional internal endoderm also referred to as the entoderm, endomesoderm, or gastrodermis as well as an outer ectoderm. On one extremity of the body are two crowns of tentacles that surround the mouth and a so-called physa on the opposite extremity. Food caught by the tentacles is ingested via a muscular and neuronal rich pharynx, and digested within the body cavity. While most of the digestive enzymes are secreted by the mesenteries that also store nutrients (Steinmetz et al., 2017), these internal structures play another crucial role as they also harbour the male or female gonads used for sexual reproduction.
Nematostella as a laboratory model
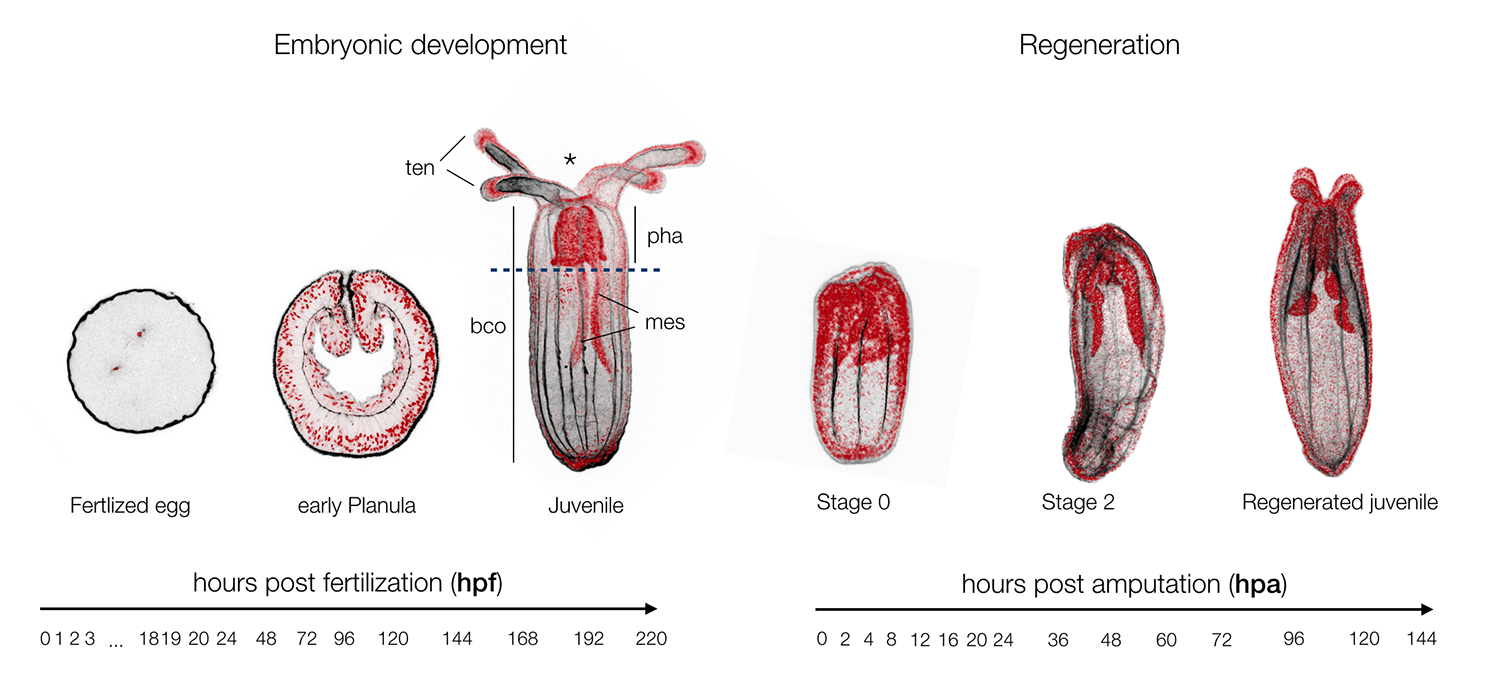
Nematostella is a cnidarian research model that was originally targeted by EvoDevo (Evolutionary & Developmental Biology) researchers looking to identify a cnidarian animal to which the development of bilaterians (insects, worms, echinoderms, vertebrates, etc.) could be compared. Studies in Nematostella have accomplished this goal and informed our understanding of the evolution of key bilaterian features. However, Nematostella is now going beyond its intended utility with potential as a model to better understand other areas such EcoDevo (Ecology & Developmental Biology) as well as regenerative biology. The latter is of particular interest as this research model has the potential to distinguish itself as a model to investigate the relationship between development and regeneration. To what extend regeneration recapitulates embryonic development is a historical question (Morgan, 1901), as both developmental trajectories lead to the same final functional organism. Being able to directly investigate and compare embryonic development and regeneration in the same species is critical to better understand the similarities of these two processes and in particular to highlight regeneration specific genes or elements (Layden et al., 2016; Warner et al., 2018).
 In 2007, the genome of Nematostella was released. Based on early comparisons between protostome and deuterostome genomes, it was believed that the cni- darian genome would be relatively simple (e.g., lacking many genes or gene families present in bilaterians). Surprisingly, the Nematostella genome is incredibly complex. Nearly all of the gene families present in bilaterians were found in Nematostella. Of note, is a nearly complete catalog of the Wnt ligands present in deuterostome bilaterians (REF 22,23). In addition, there was considerable syntenic conservation between stretches of cnidarian and vertebrate genomes and high conservation of intron-exon boundaries with vertebrate genomes (REF 23). All told at the genomic level, vertebrates and Nematostella are more similar to each other than either group is to ecdysozoan model animals (Drosophila and nematodes, REF 24). Thus, the first insight about evolution of bilaterian complexity was that complex molecular architecture predated the emergence of bilaterians and that there is no simple correlation between organismal and genomic complexity. The quest for such a correlation is still attracting attention and has driven the adaptation of new experimental tools for Nematostella: micro-RNAs and other smallRNAs have been isolated sys- tematically (REF 25,26). ChIPseq has been used to identify epigenetic marks and predict regulatory elements (REF 27) and improved transcriptome resources can provide insight into the prevalence of alternative splicing and long non-coding RNAs REF 28–30).
In 2007, the genome of Nematostella was released. Based on early comparisons between protostome and deuterostome genomes, it was believed that the cni- darian genome would be relatively simple (e.g., lacking many genes or gene families present in bilaterians). Surprisingly, the Nematostella genome is incredibly complex. Nearly all of the gene families present in bilaterians were found in Nematostella. Of note, is a nearly complete catalog of the Wnt ligands present in deuterostome bilaterians (REF 22,23). In addition, there was considerable syntenic conservation between stretches of cnidarian and vertebrate genomes and high conservation of intron-exon boundaries with vertebrate genomes (REF 23). All told at the genomic level, vertebrates and Nematostella are more similar to each other than either group is to ecdysozoan model animals (Drosophila and nematodes, REF 24). Thus, the first insight about evolution of bilaterian complexity was that complex molecular architecture predated the emergence of bilaterians and that there is no simple correlation between organismal and genomic complexity. The quest for such a correlation is still attracting attention and has driven the adaptation of new experimental tools for Nematostella: micro-RNAs and other smallRNAs have been isolated sys- tematically (REF 25,26). ChIPseq has been used to identify epigenetic marks and predict regulatory elements (REF 27) and improved transcriptome resources can provide insight into the prevalence of alternative splicing and long non-coding RNAs REF 28–30).
Because of the sequencing of the Nematostella genome, tool development has occurred on a relatively rapid scale providing new investigators an array of mechanisms for incorporating Nematostella into their research. Table 1 highlights all the tools and resources available for Nematostella and provides references and hyperlinks to resource pages for protocols and genomic tools. First, the sequenced annotated genome offers the ability to rapidly identify gene homologs (Ref 23). Although a second generation genome is still in the works, transcriptome studies provide refined gene models (Ref 28,29). Tools for visualizing gene expression and protein localization using transgenics, mRNA in situ hybridization, injection of mRNA fusion constructs, and antibody staining have been developed (REF 12,31,38,42). The ability to knockdown gene function using morpholinos or knockout gene function using CRISPR/Cas9 or TALEN-Fok approaches is now available (REF 35–38). Overexpression via mRNA injection allows for misexpression in early embryos (REF 19,30,38,39). Recently, a heat-shock responsive promoter was identified paving the way for more complex inducible constructs and the ability to test gain of function phenotypes at later stages of development (REF 35). Technology improving conditional gene regulation would strengthen Nematostella’s utility as a model system. Protocols for ChIPseq and RNAseq have also been developed (REF 27–29). To date Nematostella has proven amenable for the adaption of methods deployed in other animal systems to disrupt gene function and analyze phenotypes in the sea anemone, and with advances like CRISPR/Cas9 technology it seems reasonable that tool development in Nematostella will continue to be relatively straight-forward.
Nematostella vectensis species ID
Common name: Starlet sea anemone
Taxon: Cnidaria (Class: Anthozoa, Order: Actinaria , Family Edwardiidae)
Developmental mode: Indirect
Life cycle: Benthic-lecithotrophic
Egg type: Telolecithal
Cleavage type: Holoblastic
Gastrulation type: Invagination
Larval type: Planula
Cited References:
Hand, C., & Uhlinger, K. R. (1994). The unique, widely distributed, estuarine sea anemone, Nematostella vectensis, Stephenson: A review, new facts, and questions. Estuaries, 17(2), 501. http://doi.org/10.2307/1352679
Reitzel, A. M., Herrera, S., Layden, M. J., Martindale, M. Q., & Shank, T. M. (2013). Going where traditional markers have not gone before: utility of and promise for RAD sequencing in marine invertebrate phylogeography and population genomics. Molecular Ecology, n/a–n/a. http://doi.org/10.1111/mec.12228
Sheader, M., Suwailem, A.M. & Rowe, G.A., 1997. The anemone, Nematostella vectensis, in Britain: considerations for conservation management. Aquatic Conservation: Marine and Freshwater Ecosystems, 7, 13-25.
Steinmetz, P. R. H., Aman, A., Kraus, J. E. M., & Technau, U. (2017). Gut-like ectodermal tissue in a sea anemone challenges germ layer homology. Nature Ecology & Evolution, 1(10), 1–8. http://doi.org/10.1038/s41559-017-0285-5
Stephenson, T.A., 1935. The British Sea Anemones, vol. 2. London: Ray Society.
Williams, R.B., 1976. Conservation of the sea anemone Nematostella vectensis in Norfolk, England, and its worldwide distribution. Transactions of the Norfolk and Norwich Naturalists Society, 23, 257-266.






